Determining the virulence of Staphylococcus aureus
1. February 2015 - 31. January 2020
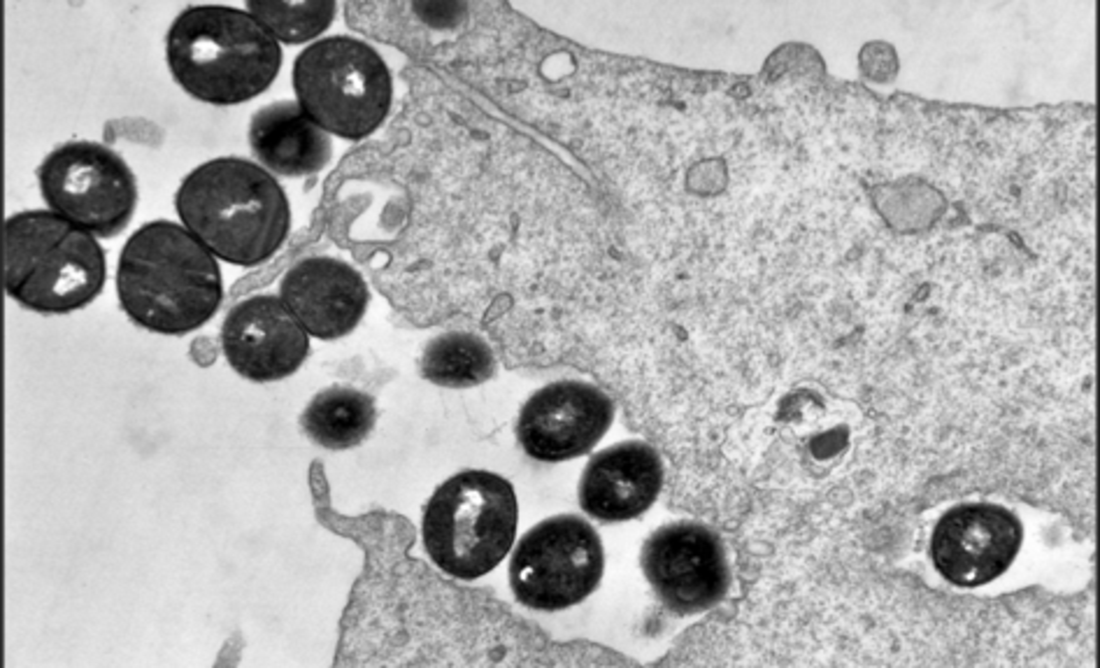
In this project, the importance of S. aureus (methicillin-resistant MRSA and methicillin-sensitive MSSA) detection in the respiratory system will be analyzed for the development of pneumonia in immunosuppressed patients. S. aureus is a facultative pathogen that colonises the nasopharynx of many healthy patients. However, colonisation is also often the starting point when an infection does occur. S. aureus can infect almost any organ in the body and this can cause very severe invasive infections such as pneumonia. Immunosuppressed patients are particularly vulnerable due to their limited defense mechanisms. To trigger infections in the body, S. aureus has a variety of virulence factors whose expression is complexly regulated and can vary greatly in different strains.
In the subproject, relevant S. aureus virulence factors that are key contributors to the development of pneumonia will be identified. Studies on the virulence profile of clinical isolates will form the basis for the development of an ELISA, PCR or microarray-based method of analysis which enables a distinction between colonising and infecting strains. This is of great importance when deciding on the initiation of antibiotic therapy, since only invasive S. aureus isolates require therapy and the importance of a positive MRSA/MSSA screening result from the nose and throat of a pneumonia patient is still unclear. Particularly in the detection of MRSA, an expensive and onerous antibiotic therapy (e.g. linezolid) is often carried out without the ability to specifically clarify the need for it beforehand.
What makes this project special is that clinical S. aureus isolates are collected from the upper and lower respiratory tract and, if applicable, recovered blood culture and pleural effusion from patients and tested for clonality and virulence. The results will be correlated with clinical data and used to characterise the strains, which takes into account the species-specific effect of many S. aureus toxins, which can sometimes be tested only in limited ways in animal models. Crucial virulence markers/virulence profiles will be checked in order to develop a diagnostic analysis method for the differentiation of colonisation and infection. With the new high-throughput methods such as next-generation sequencing (NGS) and mass spectrometry, it is even possible to test S. aureus isolates not only for the expression of known virulence factors, but also to identify factors that have not been previously described and characterised.
This research project is part of the campus project "Pneumonia in Immunosuppression"
Project duration
01.02.2015 – 31.01.2020
Project coordination
University Hospital Jena